The Goal of the SFB 1381
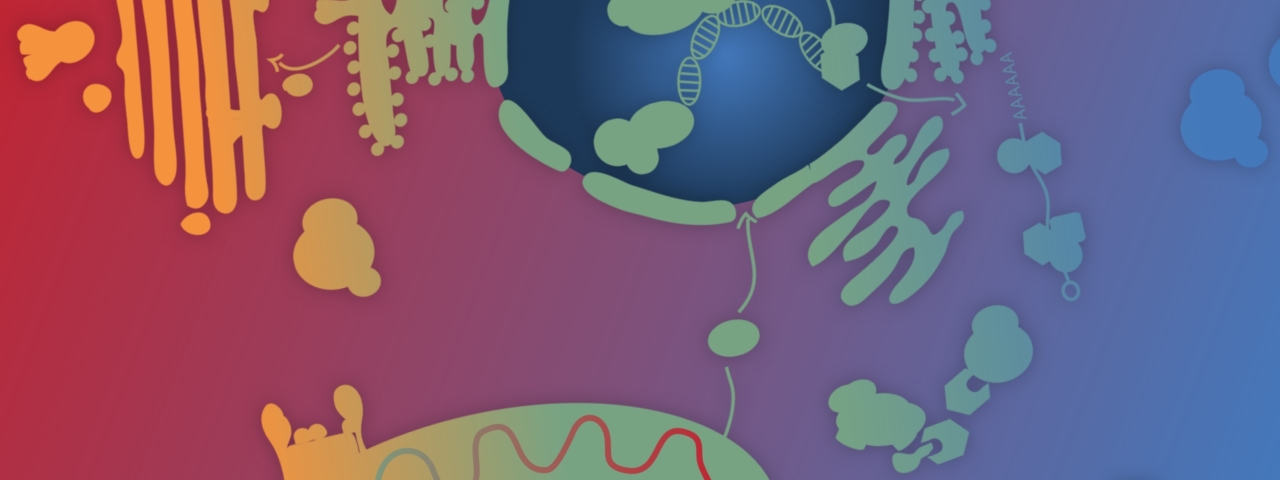
The SFB 1381 investigates how different proteins are dynamically assembled into complex multimers, the so-called protein machineries, which play a central role e.g. in the energy metabolism of the cell, the replication, repair, and transcription of DNA, the folding and degradation of proteins, as well as the intra- and intercellular communication and transport processes. The focus of the SFB 1381 lies on the organization of these protein machineries in modular units, the regulation of their assembly and disassembly, and the impact of these processes on cellular functions.
The aim of the SFB 1381 is to define and understand the dynamic processes of the assembly and organization of cellular protein machineries, and to gain insight into the fundamental processes in a living cell.
Imagefilm of SFB 1381. Video Jürgen Gocke. Also available on Youtube
Upcoming Events
News
Selected Publications
Heinen, S., Tiku, V., Grevel, A., Hugenroth, M., Ellenrieder, L., Steymans, I., Licheva, M., Bonini, S., Oeljeklaus, S., Okon, N., Lambertz, J., Poppe, S., Song, J., Fester, L., Meisinger, C., Warscheid, B., Eckhardt, M., Wiedemann, N., Winter, D., Kraft, C., den Brave, F., Bohnert, M., Pfanner, N. and Becker, T. (2026) Mitochondria contact lipid droplets through the mitochondrial import complex binding to lipid metabolism enzyme Ayr1. Nat. Cell Biol. online ahead of print
Zhang, Y., Grundmann, L., Vollmar, L., Schimpf, J., Hubscher, V., Areeb, M., Grishkovskaya, I., Moddemann, A., Werner, K., Hugel, T., Haselbach, D. and Rospert, S. (2026) The cotranslational cycle of the ribosome-bound Hsp70 homolog Ssb. Nat. Commun. 17, 961
Scheinost, L., Ludwig, C., Höfflin, N., Taskin, A.A., Marada, A., Vögtle, F.N., Meisinger, C. and Köhn, M. (2026) Synthetic trap-peptides identify a TOM complex phosphatase – PP2A dephosphorylates Tom6. FEBS J. 293, 271-294
Samir, S., Doello, S., Enkerlin, A.M., Zimmer, E., Haffner, M., Muller, T., Dengler, L., Lambidis, S.P., Sivabalasarma, S., Albers, S.V. and Selim, K.A. (2025) The second messenger c-di-AMP controls natural competence via ComFB signaling protein. Cell Discov. 11, 65
Sindram, E., Deau, M.C., Ligeon, L.A., Sanchez-Martin, P., Nestel, S., Jung, S., Ruf, S., Mishra, P., Proietti, M., Günther, S., Thedieck, K., Roussa, E., Rambold, A., Münz, C., Kraft, C., Grimbacher, B. and Gámez-Díaz, L. (2025) LRBA deficiency impairs autophagy and contributes to enhanced antigen presentation and T-cell dysregulation. EMBO Rep. 1-32-32